|
|
| (67 intermediate revisions by 4 users not shown) |
| Line 1: |
Line 1: |
| − | ==Research Summary==
| + | [[File:research_vision_20230823.png|center|1200px]] |
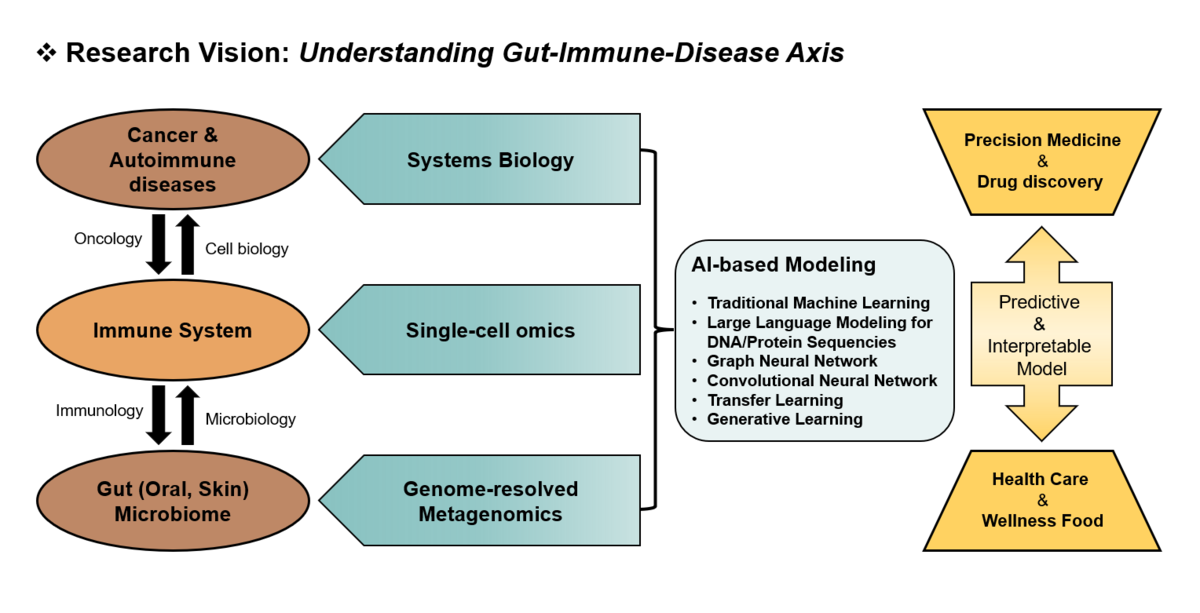
| − | The ultimate goal of biological research is to manipulate traits that are important for medicine, agriculture, and bio-industry. This challenging task first requires good understanding of association between genotype and phenotype. Because of high complexity of genotype as well as phenotype, complexity of the genotype-phenotype association could be even untouchable by combinatorial explosion of the number of possible associations. Therefore, modern genetics needs to be more systematic and predictive. Recently we proposed network-guided approach for genetics of complex traits. First, we construct probabilistic functional gene networks for cells or organisms by benchmarking and integrating heterogeneous multi-omics data that are in general publicly available. Then, using guilt-by-association, and other algorithms of network propagation of known biological information, we predict gene functions, phenotypic effect of loss-of-function, and epistatic interaction. The information can contribute to reconstruction of map between genotype and phenotype. The network-guided genetics method has been effectively applied for various organisms; from simple microbe yeast, to multicellular animal C. elegans, to the reference plant Arabidopsis, to the reference crop rice, and to the human.
| + | |
| − | | + | |
| − | ==Research Philosophy==
| + | |
| − | '''4P Research (Play, Ponder, Pride, Provide)'''
| + | |
| − | *'''Play''': Your lab must be your playground.
| + | |
| − | *'''Ponder''': More thinking, Less labor.
| + | |
| − | *'''Pride''': Be proud of yourself and your work.
| + | |
| − | *'''Provide''': People supported your work, thus you must return your discovery to people.
| + | |
| − | | + | |
| − | '''Good science must make scientists happy.'''
| + | |
| − | | + | |
| − | '''The more learn, the less know.'''
| + | |
| − | | + | |
| − | ==Research Highlight==
| + | |
| − | *[[media:research_highlight_001.pdf|Networking an organism]]
| + | |
| − | *[[media:research_highlight_002.pdf|Predicting phenotypic effects of gene perturbations in ''C. elegans'' using an integrated network model]]
| + | |
| − | *[[media:research_highlight_003.pdf|Wormnet: a crystal ball for ''Caenorhabditis elegans'']]
| + | |
| − | *[[media:research_highlight_004.pdf|A network of interactors]]
| + | |
| − | *[[media:Gallery_NGcover_SmallVer.jpg|Network perturbation predicts phenotype]]
| + | |
| − | | + | |
| − | ==Collaborators==
| + | |
| − | *[http://www.marcottelab.org/index.php/Main_Page Edward Marcotte, University of Texas at Austin, USA]
| + | |
| − | *[http://www.fraserlab.org/ Andrew Fraser, University of Toronto, Canada]
| + | |
| − | *[http://www.crg.es/ben_lehner Ben Lehner, Systems Biology Unit, EMBL-CRG, Spain]
| + | |
| − | *[http://dpb.carnegiescience.edu/labs/rhee-lab Sue Rhee, Carnegie Institution of Science, USA]
| + | |
| − | *[http://indica.ucdavis.edu/ Pamela Ronald, University of California at Davis, USA]
| + | |
| − | *[http://www.sanger.ac.uk/research/faculty/mhurles/ Matthew Hurles, Sanger Institute, UK]
| + | |
| − | *[http://www.biology.duke.edu/benfeylab/index.htm Philip Benfey, Duke University, USA]
| + | |
| − | *Sangsun Yoon, Yonsei Medical School, Korea
| + | |
| − | *[http://www.chamc.co.kr/professor/drleedr/ Dongryul Lee, Cha Medical School, Korea]
| + | |
| − | *[http://www.bahnlab.com/ Yongsun Bahn, Yonsei University, Korea]
| + | |
| − | *[http://web.yonsei.ac.kr/labsi/ Sangjun Ha, Yonsei University, Korea]
| + | |